Blog Archives
Amber and Alissa to present at Student Seminar on Thursday (16.December) at 12:30 in 2-501 BSB
Amber and Alissa will be presenting Student Seminar this week in 2-501 BSB at 12:30. I have asked the speakers to provide a brief overview for their talks. Hope to see you there!
Background for Amber’s talk
Understanding the Role of Topoisomerase 2 in Chromosome Interactions
Homologous chromosomes display associations in many organisms. Many of these interactions lead to changes in gene expression, as exemplified by processes of transvection, mammalian X-inactivation, and imprinting. Drosophila serves as an excellent model to study pairing interactions because homologous chromosomes are fully aligned in all cells. One gene required for these homolog interactions is Topoisomerase II (Top2; Williams et al. 2007). Top2 encodes an ATP dependent homodimeric enzyme that generates double stranded breaks (DSBs) to change DNA topology.
The catalytic cycle of Top2 depends on a number of functional domains. The ATPase domain is a member of the GHKL superfamily (Gyrase, Hsp90, CheA-type histidine Kinase, and MutL) that is common to all type II topoisomerases [Schoeffler and Berger 2008]. This domain forms a clamp structure that captures a DNA segment (often referred to as the G segment). Adjacent to the ATPase domain is a transducer domain that signals to the ATPase domain that the G segment is bound and cut. Top2 cleavage requires an acidic triad of amino acids within the TOPRIM domain that binds Mg+2 and interacts with the active site tyrosine of the winged helix domain (WHD) of the partner subunit. DNA cleavage occurs when the active site tyrosine covalently attaches to the 5’ end. As such, DNA cleavage by Top2 does not generate unprotected DNA ends, preventing activation of a DNA checkpoint. DNA binding is aided by the tower domain that lies next to the WHD. The C-terminus continues in a coiled-coil domain important for DNA segment release (known as T segment release).
Top2 is important in many biological processes, including DNA replication, recombination, chromosome condensation, transcription, and the segregation of sister chromatids in mitosis and meiosis [reviewed in Nittiss 2009]. Top2 is a target for many chemotherapeutic drugs because enzyme-mediated DNA damage causes cells to undergo apoptosis. To date, published Top2 studies in Drosophila have used RNAi or chemical inhibitors to understand the function of the enzyme in vivo. To expand our understanding of the role of Top2 in chromosome pairing, we conducted a genetic screen to isolate germ line mutations in Top2. Fourteen recessive lethal alleles were identified from a screen of three thousand chromosomes. Molecular analyses of these alleles uncovered single or multiple base pair substitutions within each mutant line. These included changes that generated premature stop codons or missense mutations within previously identified functional domains. Even though flies carrying each missense allele in trans to a deficiency produce inviability, hetero-allelic combinations of several missense alleles produce live flies. Current studies are directed at using the viable hetero-allelic combinations to examine chromosome structure and function during development. These data suggest that the collection of Top2 mutations will provide a resource for understanding chromosome functions that require homolog interactions.
Background for Alissa’s talk
A NOVEL ROLE FOR A CDC42 EFFECTOR PROTEIN IN XENOPUS DEVELOPMENT AND NEUROGENESIS
Many cellular functions, such as axis determination and cell movement, are dependent on the asymmetrical distribution of proteins and RNAs. In Xenopus laevis, several maternal mRNAs essential for normal development are localized to the oocyte vegetal cortex, and our lab has used microarrays to identify numerous additional localized RNAs. In this work we characterize one of these cortex-enriched transcripts, cep4l. CEP4L belongs to a family of Cdc42 effector proteins (CEPs) that bind Cdc42 and related small GTPases, which can regulate the actin cytoskeleton. Using in situ hybridization we found that cep4l is expressed in the oocyte vegetal cortex and throughout embryonic development. The embryonic expression pattern includes migratory cell populations during gastrulation and neurulation, and neural regions in older embryos. To begin to ascertain the function of cep4l in development we misexpressed cep4l RNA in embryos. We found that this overexpression causes convergent extension defects and induces ectopic neuronal marker expression, indicating a role in neurogenesis. Structure-function analysis using deletion mutant constructs show a role for the CRIB domain, a conserved GTPase binding domain. Experiments to identify upstream and downstream pathways indicate a potential role for FGF. Co-expression studies with FGF8a, which also induces ectopic neurons, demonstrate an enhancement of the neurogenic phenotype. We also present loss of function data showing a role for cep4l in normal axial and nervous system development, as well as a requirement for FGF8a-induced neurons. Although the roles of small GTPases in cell migration and adhesion are well-characterized, our results suggest novel roles for these proteins and their effectors in neurogenesis and early development.
Alissa and Janine to present st Student Seminar Thursday (18.Feb) at noon in 2-501 BSB
Alissa and Janine will be presenting Student Seminar this week in 2-501 BSB at noon. I have asked the speakers to provide a brief overview for their talks. Hope to see you there!
Background for Alissa’s talk
THE ROLE OF A NOVEL CDC42 EFFECTOR PROTEIN IN XENOPUS DEVELOPMENT AND NEUROGENESIS
Many cellular functions, such as axis determination and cell movement, are dependent on the asymmetrical distribution of proteins and RNAs. In Xenopus laevis, several maternal mRNAs essential for normal development are localized to the oocyte vegetal cortex, and our lab has used microarrays to identify numerous additional localized RNAs. In this work we characterize one of these cortex-enriched transcripts, cep4l. CEP4L belongs to a family of Cdc42 effector proteins (CEPs) that bind Cdc42 and related small GTPases, which can regulate the actin cytoskeleton. Using in situ hybridization we found that cep4l is expressed in the oocyte vegetal cortex and throughout embryonic development. The embryonic expression pattern includes migratory cell populations during gastrulation and neurulation, and neural regions in older embryos. To begin to ascertain the function of cep4l in development we misexpressed cep4l RNA in embryos. We found that this overexpression causes convergent extension defects, cell disaggregation, and induces ectopic neural and neuronal marker expression, indicating a potential role in neurogenesis. Structure-function analyses using deletion mutant constructs show a role for the CRIB domain, a conserved GTPase binding domain. We also present loss of function data showing a role for cep4l in normal axial and nervous system development. Although the roles of small GTPases in cell migration and adhesion are well-characterized, our results suggest novel roles for these proteins and their effectors in neurogenesis and early development.
Background for Janine’s talk
DYT1 (OMIM) is the most common form of inhe rited dystonia (OMIM), a disabling neurologically based movement disorder. This incurable disease is caused by the deletion of a glutamic acid residue in the protein torsinA (torA(ΔE)). Because this mutation is found in nearly all DYT1 patients and that torA(ΔE) is believed to act in a dominant negative manner over torA(WT), allele-specific silencing of torA(ΔE) may be a potential therapeutic strategy for DYT1. My research involves developing a viral mediated RNAi therapy and test both its safety and effectiveness in DYT1 murine models. Optimization of therapeutic vectors and plans for ongoing and future experiments will be discussed.
rited dystonia (OMIM), a disabling neurologically based movement disorder. This incurable disease is caused by the deletion of a glutamic acid residue in the protein torsinA (torA(ΔE)). Because this mutation is found in nearly all DYT1 patients and that torA(ΔE) is believed to act in a dominant negative manner over torA(WT), allele-specific silencing of torA(ΔE) may be a potential therapeutic strategy for DYT1. My research involves developing a viral mediated RNAi therapy and test both its safety and effectiveness in DYT1 murine models. Optimization of therapeutic vectors and plans for ongoing and future experiments will be discussed.
New GSS respresentative
Thank you Tara Maga for serving as our GSS representative for the past year.
Alissa Hulstrand has agreed to serve as Genetics representative to the Graduate Student Senate.
Ahmed and Alissa to Present @ Student Seminar on 5.14.09
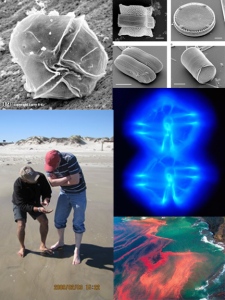
Click for larger image. Up-left + Mid-right: alexandrium1 and alexandrium2: Toxic dinoflagellate Alexandrium. Up-right: diatoms: Diatoms cells (J. Bradbury; http://dx.doi.org/10.1371/journal.pbio.0020306). Bottom-left: teaching_moment: Red tide functional genomics group meeting at Port Aransas, Texas. Bottom-right: red_tide: A non-toxic red tide bloom of Noctiluca scintillans in New Zealand. (M. Godfrey; http://www.whoi.edu/redtide/)
Ahmed Moustafa and Alissa Hulstrand will be presenting @ Student Seminar this week on the eastside of campus in 106 BBE @ noon. As usual, I have asked the speakers to provide some background information for their talks. Hope to see you all there.
Background for Ahmed’s Talk
Abstract:
Diatoms and other chromalveolates are among the dominant phytoplankton in the world’s oceans. Endosymbiosis was essential to the success of chromalveolates and it appears the ancestral plastid in this group of red algal originated via a single, ancient secondary endosymbiosis. However, recent analyses have turned up a handful of nuclear genes in chromalveolates that are of green algal derivation. Using a genome-wide approach to estimate the “green” contribution to diatoms, we identified >1,700 green gene transfers, comprising 16% of the diatom nuclear coding potential. These genes were likely introduced into diatoms and other chromalveolates from a cryptic endosymbiont related to prasinophyte-like green algae. Chromalveolates appear to have recruited genes from the two major existing algal groups to forge a highly successful, species-rich protist lineage.
Research:
Focuses on the functional genomics of harmful algae (Red Tide) and evolutionary genomics of marine microalgae. Working on characterizing and determining the origin and assembly of the saxitoxin gene cluster in cyanobacteria. Also, profiling global gene expression patterns in dinoflagellates and the impact of bacterial interaction on the transcriptional regulation in dinoflagellates.
Background for Alissa’s Talk
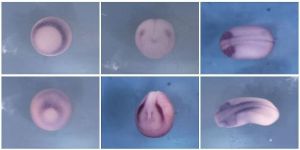 Many cellular functions, such as axis determination and cell movement, are dependent on the asymmetrical distribution of proteins and RNAs. In Xenopus laevis, several maternal mRNAs essential for normal development are localized to the oocyte vegetal cortex, and our lab has used microarrays to identify numerous additional localized RNAs. In this work we characterize one of these cortex-enriched transcripts, cep4l. CEP4L belongs to a family of Cdc42 effector proteins (CEPs) that bind Cdc42 and related small GTPases, which can regulate the actin cytoskeleton. Using in situ hybridization we found that cep4l is expressed in the oocyte vegetal cortex and throughout embryonic development. The embryonic expression pattern includes migratory cell populations during gastrulation and neurulation, and neural regions in older embryos. To begin to ascertain the function of cep4l in development we misexpressed cep4l RNA in embryos. We found that this overexpression causes convergent extension defects, cell disaggregation, and induces ectopic neural and neuronal marker expression, indicating a potential role in neurogenesis. Structure-function analyses using deletion mutant constructs show a role for the CRIB domain, a conserved GTPase binding domain. We also present loss of function data showing a role for cep4l in normal axial and nervous system development. Although the roles of small GTPases in cell migration and adhesion are well-characterized, our results suggest novel roles for these proteins and their effectors in neurogenesis and early development.
Many cellular functions, such as axis determination and cell movement, are dependent on the asymmetrical distribution of proteins and RNAs. In Xenopus laevis, several maternal mRNAs essential for normal development are localized to the oocyte vegetal cortex, and our lab has used microarrays to identify numerous additional localized RNAs. In this work we characterize one of these cortex-enriched transcripts, cep4l. CEP4L belongs to a family of Cdc42 effector proteins (CEPs) that bind Cdc42 and related small GTPases, which can regulate the actin cytoskeleton. Using in situ hybridization we found that cep4l is expressed in the oocyte vegetal cortex and throughout embryonic development. The embryonic expression pattern includes migratory cell populations during gastrulation and neurulation, and neural regions in older embryos. To begin to ascertain the function of cep4l in development we misexpressed cep4l RNA in embryos. We found that this overexpression causes convergent extension defects, cell disaggregation, and induces ectopic neural and neuronal marker expression, indicating a potential role in neurogenesis. Structure-function analyses using deletion mutant constructs show a role for the CRIB domain, a conserved GTPase binding domain. We also present loss of function data showing a role for cep4l in normal axial and nervous system development. Although the roles of small GTPases in cell migration and adhesion are well-characterized, our results suggest novel roles for these proteins and their effectors in neurogenesis and early development.